P03: Computational models of the epigenetic potential in memory
(PI: Nikolaus Fortelny)
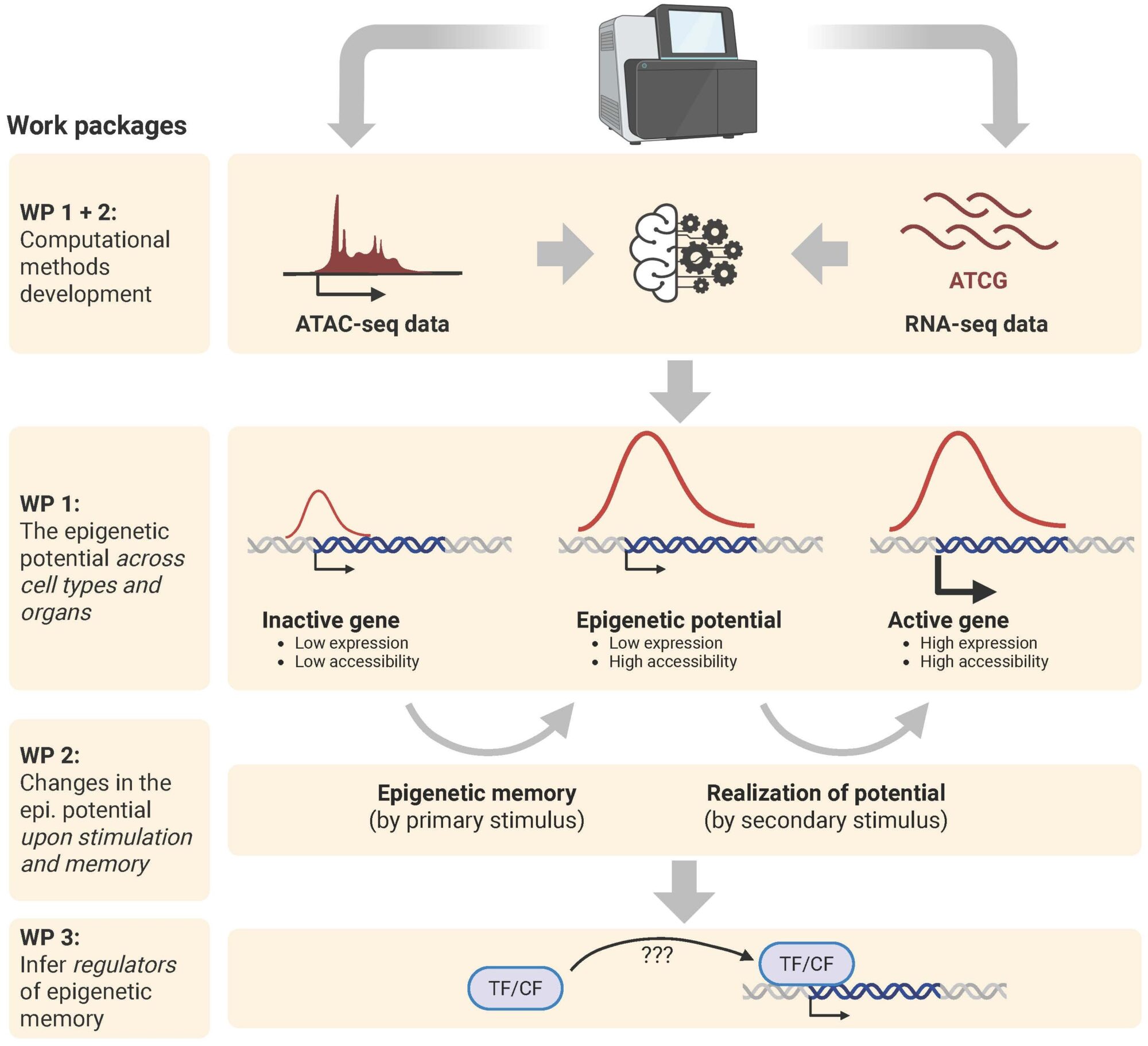
The project will computationally analyze how inflammatory memory is epigenetically encoded. We previously pioneered the concept of the “epigenetic potential”, i.e. genes that are lowly expressed but have open chromatin (Krausgruber and Fortelny, Nature, 2020). These genes have the potential to be rapidly activated upon stimulation.
The epigenetic potential has been analyzed in structural cells but a broad characterization across different cell types is currently lacking. Importantly, the epigenetic potential of a cell can change between conditions such as inflammation but computational methods to analyze such changes jointly across omics are currently not available.
In this project, we will develop novel computational approaches (using statistics as well as AI/ML) to integrate and directly compare the epigenome and transcriptome at baseline states and upon stimulation. We will then compare the epigenetic potential across cell types, organs, and inflammatory conditions using public data and those generated in EpiFlaMe. We will predict molecular mechanisms that regulate observed epigenetic changes. The project will entail collaborations with all other EpiFlaMe subprojects, analyzing the generated data and by providing predictions for experimental validation experiments.
Keywords: bioinformatics, AI/ML, epigenetic regulation





