P06: Inflammatory memory in skin and cutaneous cancer
(PI: Iris Gratz)
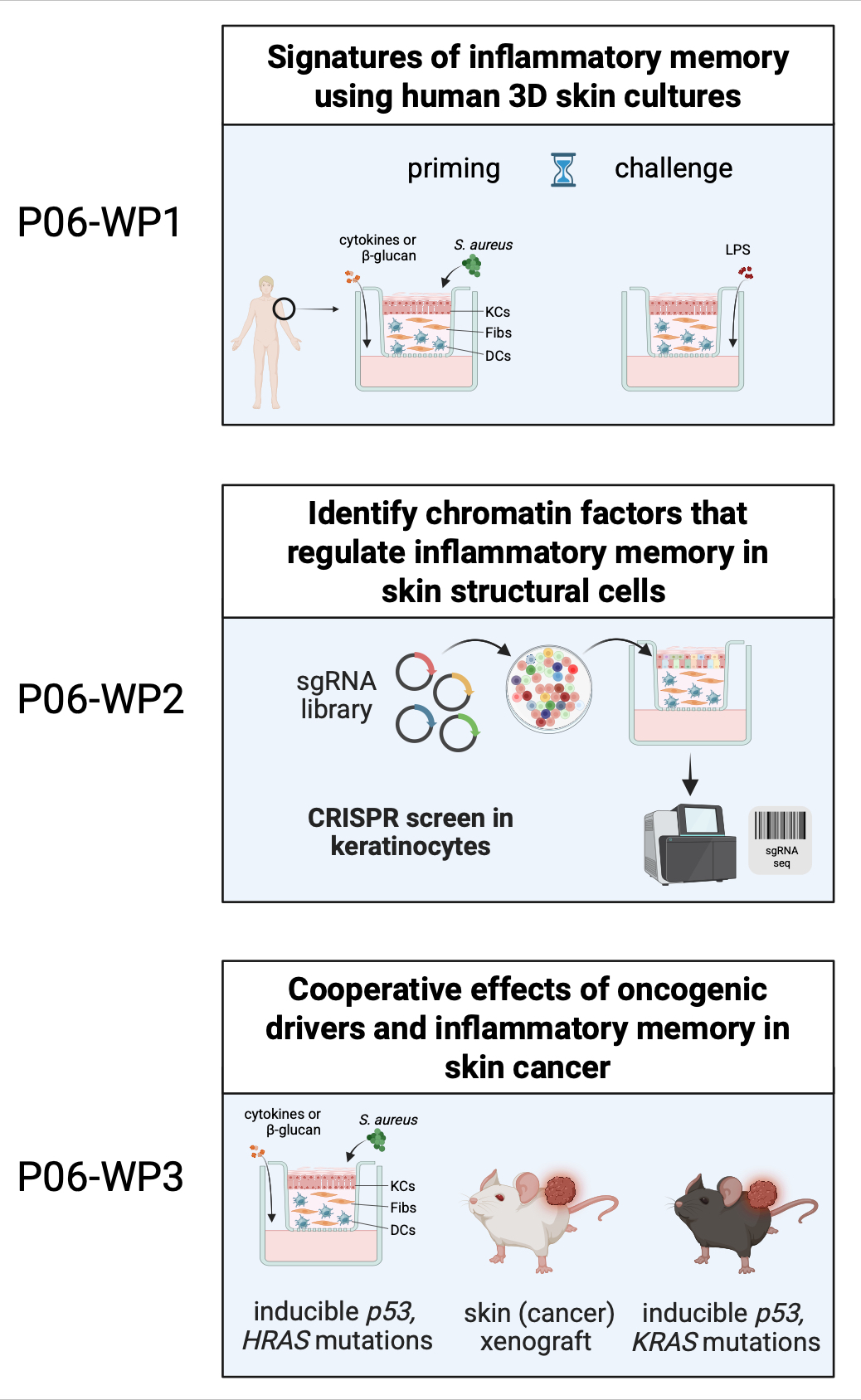
Although it is evident that skin structural cells are epigenetically modified upon inflammation, the mechanistic basis of this regulation is rather unclear. We have previously developed and optimized composite organotypic skin co-culture systems in which we found that all included cell types (keratinocytes, fibroblasts, dendritic cells) respond to stimulation (e.g., with b-glucan), which represents an exciting possibility of studying multiple cell types in a highly controlled environment. We will exploit this model to systematically map the molecular responses to b-glucan, cytokines (IL-1β, TNF, or IL-6), or infection (with S. aureus and S. epidermidis). To dissect molecular regulators of inflammatory memory, we will perform CRISPR screens in cutaneous structural cells and individually validate their function. Finally, we will elucidate the cooperative effects of inflammation and oncogene alleles (HRAS), or deficiency of the tumor suppressor gene TP53 in cancer. We will validate our findings, using our in vitro and in vivo models (including skin/tumor xenografting models). Our results will for the first time reveal cell-type- versus organ-specific inflammatory memory in skin. Crucially, we will reveal signatures unique to skin or shared with other organs and we will link those to their functional consequences in skin cancer in preclinical experiments.
Together, P06 will test the hypothesis that proinflammatory molecules and skin microbes reprogram local KCs and Fibs during inflammation, by altering their epigenetic and transcriptional landscape and their responses to subsequent stimuli, and that this influences the course of skin cancer.
Keywords: Immunology, skin, inflammation, infection





