Physiochemical Principles of Immune Interfaces
Our lab explores how the physical and chemical organization of cell surfaces governs immune recognition, communication and cytotoxicity. We investigate the nanoscale interface where immune cells meet other immune cells, tumour cells, or engineered substrates and uncover how receptor–ligand interactions, membrane mechanics and cytoskeletal dynamics together determine the outcome of an immune response.
Our work combines biophysics, molecular immunology, synthetic biology and advanced imaging with AI-assisted analysis to dissect the earliest moments of immune synapse formation. By reconstructing immune interfaces with precisely controlled reductionist systems, such as supported lipid bilayers or microfluidic devices, and by studying immune cell interactions in tissue-like environments, we bridge reductionist mechanistic insight with physiological relevance.
A key theme of the lab is understanding how ligand mobility, molecular anchoring, nanoscale organization and membrane stiffness shape receptor signalling and force transmission. We investigate how dendritic cells and tumour cells regulate these properties through cytoskeletal coupling, and how such regulation can enhance immune activation, dampen it, or enable immune evasion. Through AI-assisted analysis of high-dimensional imaging data, we map the dynamic architectures that predict T cell behaviour and cytotoxic efficiency.
This mechanistic foundation is directly relevant for the design and optimization of modern immunotherapies. Therapies such as chimeric antigen receptor T cells (CAR T) cells, Bispecific T cell engagers (BiTEs), checkpoint inhibitors, and small-molecule immunomodulators all rely on how T cells interpret the physicochemical properties of the surfaces they encounter. By defining the biophysical rules that govern synapse formation and early signalling, we aim to reveal why certain therapeutic formats succeed, why others fail and how molecular features can be tuned to improve potency and precision.
By reconstructing immune interfaces across three levels, precisely controlled reductionist systems (e.g., supported lipid bilayers (Fig 1.), microfluidic platforms), direct cell–cell interactions (Fig. 2), and cell–cell dynamics within tissue-like environments (Fig. 3), we connect quantitative mechanistic insight with physiological relevance.
Across all these efforts, our goal is to build a mechanistic, physicochemical framework of immune interfaces, revealing how immune synapses form, how signals are integrated, and how biophysical constraints influence therapeutic success. We aim to translate this understanding into improved strategies for modulating T cell function and for designing next-generation immunotherapies.
Our research focuses on:
- Biophysical regulation of immune synapse formation, including receptor–ligand dynamics, spatial patterning and signal integration.
- Cytoskeletal control of ligand presentation on dendritic cells and tumour cells, and its impact on T cell activation, suppression and immune evasion.
- Physicochemical determinants of immune recognition, such as ligand lateral mobility, mechanical resistance, molecular crowding, and force loading.
- Synthetic reconstruction of immune interfaces, using mixed-mobility lipid bilayers and engineered ligand systems to precisely tune the presentation of immune cues.
- T cell activation and cytotoxicity in physiologically relevant environments, integrating biophysical mechanisms with tissue-like models and dynamic live-cell imaging.
By uncovering the rules that govern how immune cells interpret the physicochemical properties of the surfaces they encounter, we aim to accelerate both fundamental discovery and the rational design of immune-based therapies, including more potent CAR T cells, optimized BiTEs and targeted immunomodulatory drugs.
Figure 1: Human T cells expressing an F-actin marker (cyan) were placed on a functionalized supported lipid bilayer that mimics the surface of a cancer cell. After initial spreading, the cells switch to centripetal actin flow, pulling T cell receptors (TCRs) inward toward the center of the immune synapse. © Alexander Leithner
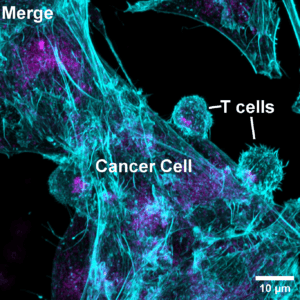
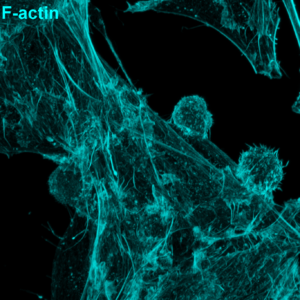
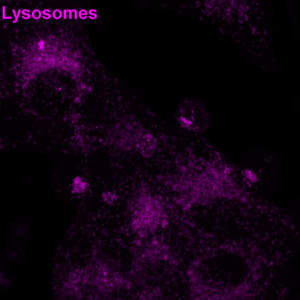
Figure 2: Human cytotoxic T cells were mixed with CD19-expressing cancer cells in the presence of a CD3–CD19 BiTE and then fixed. F-actin (cyan) and lysosomes (magenta) areshown. Notice how T cell lysosomes gather at the contact point, ready to deliver their cytotoxic payload. © Alexander Leithner
Figure 3: Murine T cells (green) and dendritic cells (red and blue) meet and interact inside the lymph node of an anesthetized mouse, visualized by two-photon intravital microscopy. © Alexander Leithner





