Computational Systems Biology
The group’s research aims to understand the function and dysfunctions of biological systems on a molecular level, focusing on the complex regulation in large networks. We rely on big biological data from multi-omics experiments (transcriptomics, proteomics, epigenomics, or others) that are often collected at single-cell or spatial resolution. The computational methods we use mirror this biological complexity, spanning advanced statistics, machine learning, and network science.
Key contributions
JAK-STAT signaling maintains homeostasis in T cells and macrophages
Fortelny*, Farlik*, et al., Nature Immunology (2024)
https://doi.org/10.1038/s41590-024-01804-1
Systematic functional screening of chromatin factors identifies strong lineage and disease dependencies in normal and malignant haematopoiesis
Lara-Astiaso*, Goñi-Salaverri*, Mendieta-Esteban* et al., Nature Genetics (2023)
https://doi.org/10.1038/s41588-023-01471-2
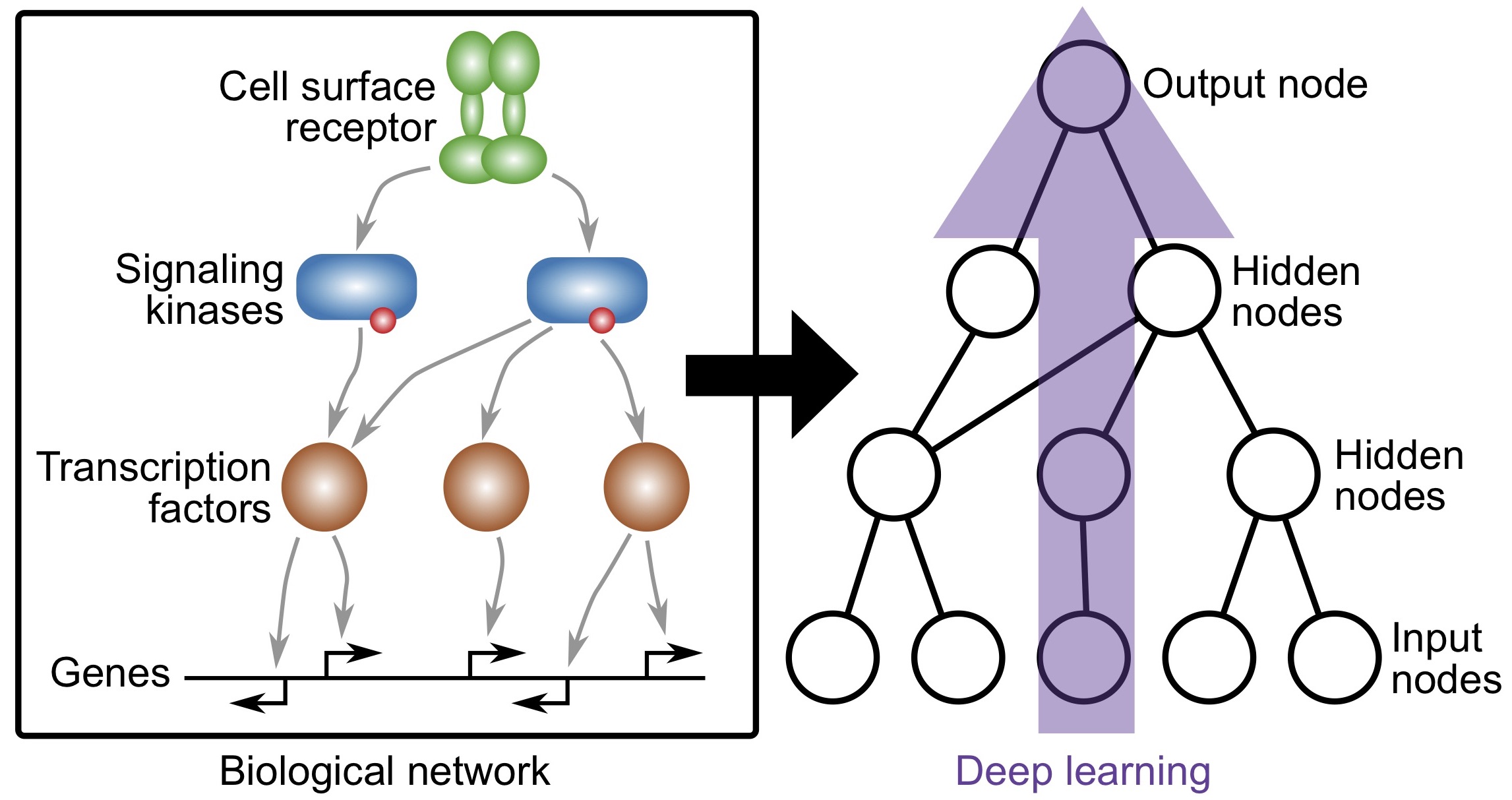
Knowledge-primed neural networks enable biologically interpretable deep learning on single-cell sequencing data
Fortelny and Bock, Genome Biology (2020)
https://doi.org/10.1186/s13059-020-02100-5
Structural cells are key regulators of organ-specific immune responses
Krausgruber* and Fortelny* et al., Nature (2020)
https://doi.org/10.1038/s41586-020-2424-4
Can we predict protein from mRNA levels?
Fortelny et al., Nature [BCA] (2017)
https://doi.org/10.1038/nature22293






