spaXio
Spatial Crosstalk in Immuno-Oncology
spaXio is an ambitious EU-funded Doctoral Network (MSCA DN) co-coordinated by Dr. Peter Krenn (Molecular Cancer Biology Lab) and Dr. Dirk Schmidt-Arras (Tumor Immunology Lab) at the University of Salzburg, Austria. This international consortium brings together leading research groups across Europe to decipher the spatial and molecular mechanisms of metastasis — how cancer cells communicate, adapt metabolically, and remodel their microenvironment to colonize distant organs. Through 14 interdisciplinary PhD projects, spaXio integrates cutting-edge cancer models, spatial multi-omics, and AI-driven data analysis to reveal the cellular and metabolic interactions driving metastatic progression. The network offers outstanding training and collaboration opportunities across Austria, Germany, France, Switzerland, and Spain. For further information please visit
spaXio Project 3: Mapping Metabolic and Cellular Interactions in Colon Cancer Metastatic Niches
Supervisors: Peter Krenn (Molecular Cancer Research Lab, Salzburg)
Co-supervisors: Manfred Claassen (Tübingen), Spencer Watson (Lausanne)
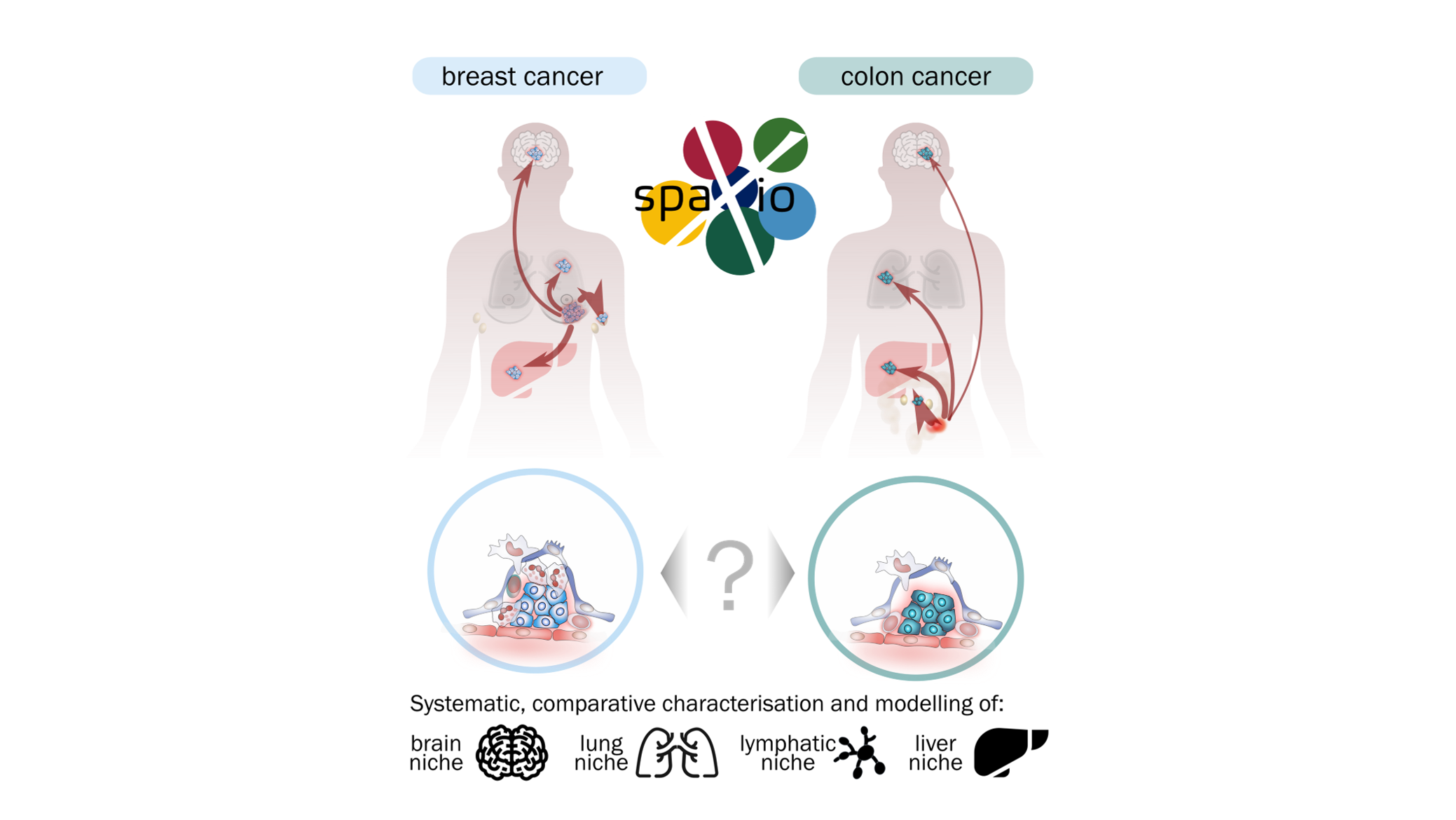
Metastasis—the spread of cancer to distant organs—is the leading cause of cancer-related mortality. Its success depends not only on the invasive properties of tumour cells but also on their ability to reprogram local tissues into supportive metastatic niches. This project investigates how metabolic adaptations and cellular interactions within these niches influence the fate of disseminated tumour cells and the establishment of secondary tumours.
The doctoral candidate of the project performed in our lab will explore metastatic niche formation using advanced experimental models that combine genetic manipulation, organoid systems, and in vivo metastasis models. Using colon organoids and orthotopic transplantation, the project will study how tumour cells interact with stromal, immune, and endothelial components during dissemination and colonisation. Organoids will be genetically engineered using CRISPR/Cas, Cre-LoxP recombination, and inducible oncogene expression systems, allowing precise spatial and temporal control of gene function and oncogenic signalling. These tools will enable functional analysis of tumour–stroma communication and metabolic reprogramming. Metastatic progression will be monitored in vivo using luciferase-based bioluminescence imaging on the AMI platform, providing non-invasive longitudinal tracking of metastatic spread. To map the molecular architecture of metastatic tissues, the project will employ spatial transcriptomics (Xenium platform), spatial metabolomics, and high-plex immunofluorescence, generating detailed molecular maps of gene expression, metabolism, and cellular organisation in lymph nodes, lung, brain, and liver. Guided by these analyses, co-culture experiments with stromal and immune cells will help functionally test the molecular mechanisms that define metastatic niches. Fluorescence and confocal live-cell microscopy will be used primarily for organoid and organoid co-culture systems, capturing dynamic cellular interactions, remodelling processes, and responses to genetic or metabolic perturbations.
By integrating genetic engineering, live imaging, and spatial multi-omics, this project aims to uncover how metabolic flexibility and intercellular communication drive metastatic niche formation. The findings will provide a mechanistic basis for identifying targetable pathways to prevent or disrupt metastatic colonisation.





